The initial phase in investigating a protein involves acquiring its precise gene sequence. Once obtained, this sequence allows for the construction of a plasmid to facilitate the expression of the desired protein.
Plasmid construction, also known as genetic engineering, begins by amplifying the selected foreign gene's DNA through Polymerase Chain Reaction (PCR). Subsequently, specific enzymes—restriction enzymes—are employed to cleave both the vector and the foreign DNA fragments. These fragments are then fused using DNA ligase and inserted into host cells. The subsequent screening process identifies the correct recombinant cloning plasmid, ensuring accurate expression of the target gene within the host cell.
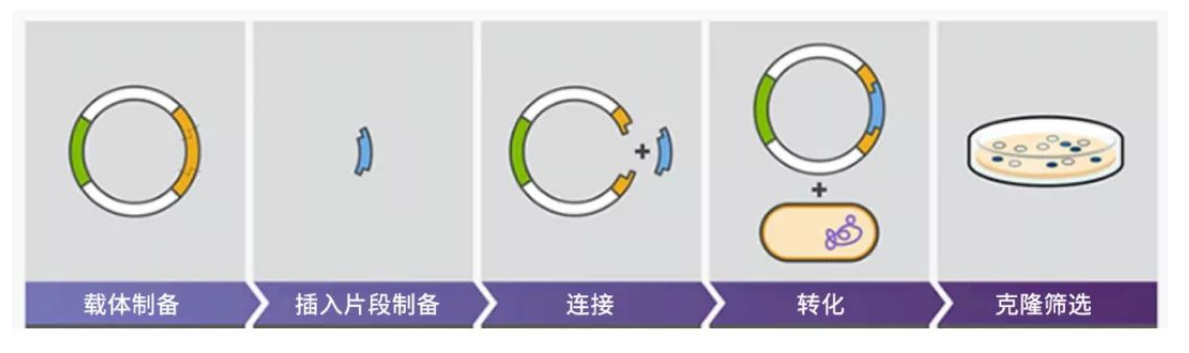
Figure 1: Schematic diagram of plasmid construction process
1. How to choose the right vector?
Vectors are usually divided into two types: cloning vectors and expression vectors.
Cloning Vectors:
Most cloning vectors are high-copy vectors, which can connect foreign genes to the plasmid of the cloning vector and introduce them into prokaryotic bacteria for large-scale replication and cloning. The main purpose is to preserve the target gene fragment.
When choosing a cloning vector, you should pay attention to:
① The ability of autonomous replication and high copy number.
② Carry selection markers that are easy to screen.
③ Contains a single recognition sequence for multiple restriction enzymes for foreign gene insertion.
④ Preference should be given to vectors smaller than 15 kilobases (kb) to facilitate their introduction into cells and enable efficient propagation.
⑤ Safety Measures: Cloning vectors must exhibit a limited host range, avoiding recombination, transfer, or the generation of harmful traits within the host. They should not propagate freely beyond the engineered host.
Expression Vectors:
Expression vectors are specially designed cloning vectors to transcribe and translate inserted foreign DNA sequences into polypeptide chains. It contains specific expression system elements, namely promoter, ribosome binding site, cloning site, and transcription termination signal.
Expression vectors can be categorized based on their expression type into four main groups:
Non-fusion expression vector, such as PKK223-3.
Secreted expression vector, such as PINIII-ompA1.
Fusion protein expression vector, such as PGEX.
Inclusion-type expression vector, such as pBV220.
Figure 2: Gene segment selection
2. Rules for primer design
When using PCR to amplify the target gene, primer design is very critical.
① The best primer length is about 18-30bp, and the commonly used length is 20-22bp.
② The Tm value of the primer should be around 60°C. The Tm value between the two primers should be kept close, and the difference should not exceed 5°C.
③ The GC content standard is usually 40%-60% or 45-55%.
④ The primer itself should not contain more than 4 consecutive complementary bases to avoid forming a hairpin structure or primer-dimer.
⑤ The 3' end of the primer should avoid continuously repeated bases, such as GGG or CCC, which will lead to mismatches. It is best for the last base to be G or C.
⑥ Adding an enzyme cutting site to the 5′ end of the primer (without affecting the specificity of amplification), different types and quantities of protective bases need to be added according to the sequence of the enzyme cutting site, usually 3 more bases are added. The base can meet the need to protect the enzyme cleavage site.
⑦ Incorporate different enzyme cleavage sites in upstream and downstream primers. Using the same enzyme cleavage site may cause the target gene fragment to link inversely, potentially impacting the gene sequence's proper expression.
3. Common Challenges in PCR Amplification
Amplifying genes via PCR is a generally straightforward process, yet it's not always guaranteed to yield a 100% success rate. Several issues can arise during amplification that affect specificity, purity, and the fidelity of the results.
a. Primer Dimer Formation and Non-Specific Bands:
The presence of primer dimers or non-specific bands of incorrect sizes can compromise amplification specificity. Address this by:
● Reducing template and primer concentrations.
● Lowering magnesium ion levels.
● Adjusting enzyme quantities.
● Increasing the annealing temperature to enhance specificity.
b. Dispersed Gel Bands:
● When gel bands appear scattered, this is often due to impure templates, imbalanced reaction components, low annealing temperatures, and excessive cycle numbers, among other factors. To rectify this, ensure:
● Purity of templates.
● Proper proportions of reaction components.
● Optimal annealing temperatures.
● Adequate cycle numbers for the specific target.
c. Challenges in Amplifying Long-Segment Genes:
● Amplifying lengthy gene segments is prone to higher rates of point mutations and mismatches. To overcome this, it's crucial to:
● Choose a polymerase known for high amplification capacity.
● Select a polymerase with high fidelity and reliability to minimize errors.
● Employ stringent quality controls throughout the amplification process.
4. Other questions
When operating the steps of enzyme digestion and ligation, it is also necessary to ensure sufficient enzyme activity qualitatively and quantitatively. For example, for double enzyme digestion, the same type of buffer should be used as much as possible. The general dosage is more than 40U units (the dosage should not exceed 1/10 of the total volume) to ensure that the enzyme activity is sufficient. The cutting process is sufficient; it can be determined according to the target fragment amount (ng) = (carrier amount (ng) × target fragment length (kb))/(carrier DNA fragment length (kb)) × molar ratio of the target fragment and vector (1:3- 1:8) Calculate the amount of target fragment and vector to add to improve the connection efficiency.
The constructed vector is put into competent cells for transformation (TOP10, DH5α, BL21, etc). The competent cells should be kept as fresh as possible when used, avoid repeated freezing and thawing, incubation on ice, and heat shock time should be strictly controlled), and the resistant cells should be, colony PCR, enzyme digestion, sequencing, and other procedures to verify whether the transformation is successful, and finally obtain the correct recombinant cloned gene fragment.
5. Selecting the Appropriate Strain and Vector (Refer to the list provided at the end of the article for specific details).
E coli strain list
| No. | strain | annotation | Resistant |
| 1 | BL21 (DE3) | most used | No |
| 2 | Rosetta (DE3) | Rare codonsAUA, AGG, AGA, CUA, CCC, GGA | Cl |
| 3 | Rosetta2 (DE3) | Rare codons AUA, AGG, AGA, CUA, CCC, GGA and CGG | Cl |
| 4 | Origami 2 (DE3) | Disulfide bonds and rare codons | StrR, Tet |
| 5 | C41 (DE3) or C43 (DE3) | hydrophobic protein | No |
| 6 | Arctic (DE3) | TPN30 & TPN60 molecular chaperones | Tet, Cam |
| 7 | Tuner (DE3) | Precisely control the expression level through IPTG,0.1mM IPTG | No |
| 8 | BL21 (DE3) pLysS | Reduce background expression of toxic proteins | Cl |
| 9 | Rosetta(DE3) pLysS | Reduce the background expression of toxic proteins, rare codonsAUA, AGG, AGA, CUA, CCC, and GGA. | Cl |
| 10 | BL21(DE3) del- slyXD | BL21(DE3)knock out slyXD | No |
| 11 | B834(DE3) | Met-deficient strain | No |
| 12 | T7 pL _ | Resistant to T1 phage infection | cl |
| 13 | BL21star (DE3) | ||
| 14 | origamiB (DE3) | disulfide bond | KanR,TetR |
| 15 | Origami 2 (DE3) pLysS | Based on Origami 2 (DE3), pLysS is added to inhibit local expression. | Cl, StrR, Tet |
| 16 | Dh5α | clonal strains | No |
| 17 | Dh5α-T1 | Resistant to T1 phage infection | No |
| 18 | Dh10bac | BacmidPreparation | Tet, gentamicin |
| 19 | BL21-Gold (DE3) | Can be used as both a protein expression strain and a plasmid cloning strain | No |
Plasmid list
| name | length | Resistance | special properties |
| PAO815 | 7709bp | Amp | yeast expression |
| wxya | 44741bp | Amp | Adenovirus expression |
| wxya | 42410 bp | Amp | Adenovirus expression |
| pAAV -MCS | 4.7kbp _ | Amp | mammalian cell expression |
| pBacPAK8 | 5.5kbp__ | Amp | Baculovirus expression |
| PBI121 | 13.0kbp_ | Kan | plant cell expression |
| pBV220 | 3665bp | Amp | prokaryotic expression |
| pCAMBIA 1300 | plant cell expression | ||
| pCAMBIA 1301 | 11837bp | Kan | plant cell expression |
| pCAT3-Basic | 4047bp | Amp | |
| pcDNA 3 | 5446bp | Amp | mammalian cellexpression |
| pcDNA 3.1(+) | 5428 bp | Amp | mammalian cell expression |
| pcDNA 3.1/ mys-HisA | 5494bp | Amp | mammalian cell expression |
| pCl -neo | 5472 bp | Amp | mammalian cell expression |
| pCMV -MCS | 4.5kbp _ | Amp | mammalian cell expression |
| pET-3a | 4640 bp | Amp | E. coli expression |
| pET-11a | 5677 bp | Amp | E. coli expression |
| pET-15b (+) | 5708 bp | Amp | E. coli expression |
| pET-20b (+) | 3716bp | Amp | E. coli expression |
| pET-22b (+) | 5493bp | Amp | E. coli expression |
| pET-23a (+) | 3666bp | Amp | E. coli expression |
| pET-23b (+) | 3665bp | Amp | E. coli expression |
| pET-23c (+) | 3664bp | Amp | E. coli expression |
| pET-23d (+) | 3663bp | Amp | E. coli expression |
| pET-28a (+) | 5369 bp | Kan | E. coli expression |
| pET-28b (+) | 5368 bp | Kan | E. coli expression |
| pET-28c (+) | 5367 bp | Kan | E. coli expression |
| pET-30a (+) | 5422 bp | Kan | E. coli expression |
| pET-30b (+) | 5421 bp | Kan | E. coli expression |
| pET-30c (+) | 5423bp | Kan | E. coli expression |
| pET-31b (+) | 5742 bp | Amp | E. coli expression |
| pET-32a (+) | 5900bp | Amp | E. coli expression |
| pET-32b (+) | 5899 bp | Amp | E. coli expression |
| pET-32c (+) | 5901 bp | Amp | E. coli expression |
| pET-39b | 6106bp | Kan | E. coli expression |
| pET-42a | 5930 bp | Kan | E. coli expression |
| pGAPZ-aA | 3147 bp | Zeo | yeast expression |
| pGBKT7 | 7.3kbp _ | Kan | yeast expression |
| pGEM3Zb | prokaryotic expression | ||
| pGEM3Zf (+) | prokaryotic expression | ||
| pGEM7Zf (+) | prokaryotic expression | ||
| pGEX-2T | 4969 bp | Amp | prokaryotic expression |
| pGEX-4T-1 | 4969 bp | Amp | prokaryotic expression |
| pGFP-N2 | 4732bp | Kan | Mammalian cells fluorescent protein expression |
| pEGFP-C1 | 4731bp | Kan | Mammalian cells fluorescent protein expression |
| pEGFP-C3 | 4727bp | Kan | Mammalian cells fluorescent protein expression |
| pEGFP-N1 | 4733bp | Kan | Mammalian cells fluorescent protein expression |
| pLEGFP-N1 | 6892 bp | Amp | Mammalian cells fluorescent protein expression |
| pGL3-Basic | 4818bp | Amp | |
| pGL36 | |||
| nnJC | 6620 bp | Amp | retroviral expression |
| nnJC | 5.6kbp _ | Amp | retroviral expression |
| nnJC | 5.9kbp _ | Amp | retroviral expression |
| ikB | 6.1kbp _ | Amp | retroviral expression |
| pMAL-p2x | 6721 bp | Amp | Prokaryotic fusion protein expression |
| pMAL-c2x | 6721 bp | Amp | Prokaryotic fusion protein expression |
| pPIC3.5K | 9004bp | Amp/Kan | yeast expression |
| pPIC9 | 8024 bp | Amp | yeast expression |
| pPIC9K | 9276 bp | Amp | yeast expression |
| pPIC aA | 3593bp | Zeo | yeast expression |
| pQpK _ | 5387bp | Amp | Mammalian cells fluorescent protein expression |
| pQE-30 | 3461bp | Amp | prokaryotic expression |
| pQE-9 | 3439bp | Amp | prokaryotic expression |
| pRevTRE | 6487bp | Amp | retroviral expression |
| pSE420L | 4617bp | Amp | |
| i _ | Amp | prokaryotic expression | |
| pTac I (BamH I) | Amp | prokaryotic expression | |
| pTAL -Luc | 4956bp | Amp | mammalian cell expression |
| pTWIN1 | 7375 bp | Amp | |
| pTXB1 | 6706bp | Amp | |
| pVAX1 | 2999 bp | Kan |




